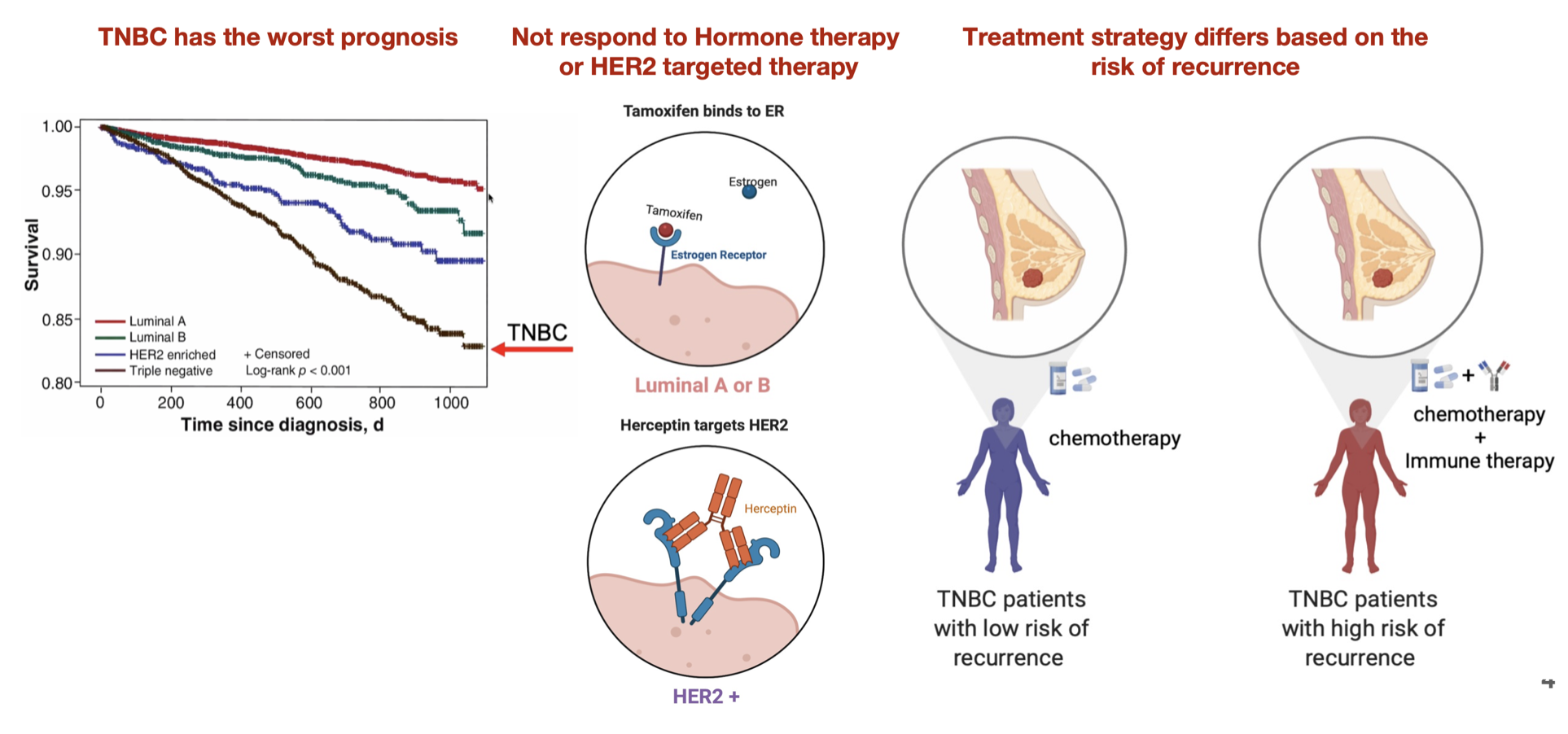
Triple Negative Breast Cancer
TNBCs are defined as lacking expression of the ER, PR, and HER2 receptors, and they tend to have a more aggressive natural history than other breast cancer subtypes.
Hiii 👋 this is Yaoyi, I'm currently a 3rd year PhD student at Baylor College of Medicine, Houston, Texas.
Before my PhD study, I finished my undergrad at Southwest University in China and received my masters degree in Biostatistics from Washington University in St. Louis, School of Medicine.
I joined Dr. Wenyi Wang's Lab at MD Anderson Cancer Center in 2021. My thesis project is about defining triple negative breast cancer patient subtypes through genomic and transcriptomic deconvolution. I will also work on developing computational methods to estimate allele-specific expression at SNV level.
Outside of the research, I enjoy many kinds of workout such as HIIT, Yoga, Pliates, and Trampoline! Cooking and Baking are my procrastinating options.

TNBCs are defined as lacking expression of the ER, PR, and HER2 receptors, and they tend to have a more aggressive natural history than other breast cancer subtypes.
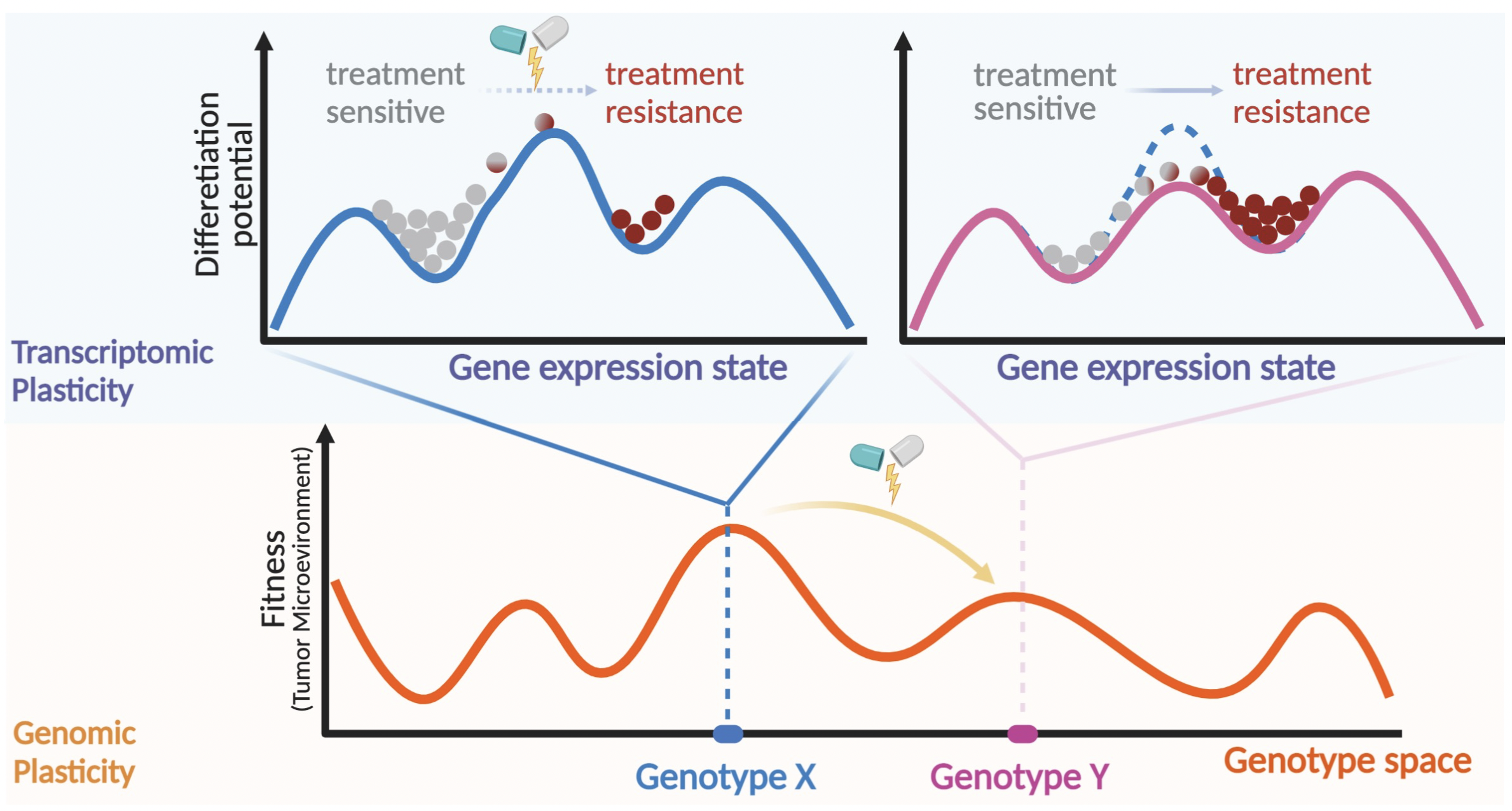
Plasticity and tumor heterogeneity present a major challenge in the clinical management of breast cancers by impacting patient’s prognosis, therapeutic response, and clinical outcomes.

In the context of cancer, allele specific expression is affected by both tumor and surrounding non-tumor cells. It is well studied that copy number alterations are highly predictive to transcript as well.

Single-cell RNA sequencing studies have suggested that total mRNA content correlates with tumor phenotypes. Technical and analytical challenges, however, have so far impeded at-scale pan-cancer examination of total mRNA content. Here we present a method to quantify tumor-specific total mRNA expression (TmS) from bulk sequencing data, taking into account tumor transcript proportion, purity and ploidy, which are estimated through transcriptomic/genomic deconvolution. We estimate and validate TmS in 6,590 patient tumors across 15 cancer types, identifying significant inter-tumor variability. Across cancers, high TmS is associated with increased risk of disease progression and death. TmS is influenced by cancer-specific patterns of gene alteration and intra-tumor genetic heterogeneity as well as by pan-cancer trends in metabolic dysregulation. Taken together, our results indicate that measuring cell-type-specific total mRNA expression in tumor cells predicts tumor phenotypes and clinical outcomes.
Alzheimer disease (AD) has substantial genetic, molecular, and cellular heterogeneity associated with its etiology. Much of our current understanding of the main AD molecular events associated with the amyloid hypothesis (APP, PSEN1 and PSEN2) and neuroimmune modulation (TREM2 and MS4A) is based on genetic studies including GWAS. However, the functional genes, downstream transcriptional ramifications, and the cell-type-specific effects of many GWAS loci remain poorly understood. Understanding these effects can point us to the cellular processes involved in AD and uncover potential therapeutic targets. We identified cell-specific expression states influenced by AD genetic factors for neurons and glia. Autosomal dominant AD (ADAD) brains exhibited unique transcriptional states in all cell types. TREM2 variant carrier brains were also enriched for specific microglia and oligodendrocyte subpopulations. Carriers of the resilience MS4A variant were enriched for an altered activated-microglia expression state. We mapped AD GWAS genes to their potential functional cell types, and some, including PLCG2 and SORL1, were expressed in a broader range of brain cell types than previously reported.